Author
Jade Ikahihifo-Bender
Mentors
Dr. Caryn Outten and Mrs. Evan Talib
Abstract
Due to its unique ability to serve as both an electron donor and acceptor, iron is utilized as a co-factor for many biological processes, including electron transfer, oxygen binding, and vitamin synthesis. Iron is also a key factor during fungal infections as the human host and invading pathogens battle over limited iron pools. The primary iron-responsive transcription factor Aft1 in the opportunistic pathogenic yeast Candida glabrata responds to iron deficiency by activating expression of iron acquisition genes. However, the mechanisms for sensing intracellular iron levels and regulating Aft1 activity in response to iron are unknown. The C. glabrata iron regulation system shares close homology to a similar system in the non-pathogenic yeast Saccharomyces cerevisiae, in which the monothiol glutaredoxins Grx3/4 and the BolA-like protein Bol2 form [2Fe-2S] binding complexes that deactivate Aft1 under iron replete conditions. To determine whether a similar mechanism controls C. glabrata Af1 activity, we sought to analyze the in vitro interactions between Grx4 and Bol2 from this yeast pathogen. For this project, we successfully subcloned the BOL2 gene from C. glabrata into Escherichia coli overexpression vectors allowing for expression of recombinant Bol2 alone or in complex with its presumed binding partner Grx4. The overexpression conditions identified here will be used in future experiments to purify and characterize the structure and iron regulation function of Bol2 alone or in a complex with Grx4.
Background
Iron is an essential nutrient that supports many cellular processes such as oxidative phosphorylation and metabolism, cellular growth and proliferation, and the transport and storage of gaseous signaling molecules.1,2 Maintenance of optimal intracellular iron levels is critical, as both iron deficiency and iron overload are detrimental to human health.1 In fact, iron deficiency is the most common nutritional deficiency in the world affecting over one billion people, and iron overload disorders are among the most common autosomal recessive disorders in Caucasian populations.1,2 Iron levels in the body are also a target for opportunistic pathogens that compete with the human host.3 An immune-compromised individual with disrupted iron sequestration would be less competitive for iron and thus subject to the virulence of an opportunistic pathogen.3 Candida glabrata, for example, is an opportunistic pathogen commonly found in healthy mucosa that is the second leading cause of candidemia, a potentially lethal fungal infection in the bloodstream.3
Our lab has used the non-pathogenic fungi Saccharomyces cerevisiae (baker’s yeast) in experiments that study the structure, function, and subcellular localization of iron binding proteins involved in iron trafficking and regulation.4-6 In this organism, the iron-responsive transcription factors Aft1 and Aft2 activate iron uptake genes in iron deplete conditions.4,7 These regulators are deactivated under iron replete conditions by binding an iron-sulfur (Fe-S) cluster.4,7 The cytosolic proteins Grx3/4 and Bol2 facilitate binding between the Fe-S cluster and Aft1/2.4
Studies utilizing this model system have delineated mechanisms that are essential to regulating iron homeostasis in a non-pathogenic yeast.4-7 The pathogenic yeast C. glabrata presumably employs a similar iron regulatory system as S. cerevisiae;8 however, little is known regarding the molecular mechanisms of iron regulation in this pathogen. Extending iron binding protein studies to C. glabrata is necessary to characterize the iron regulation pathways that are essential to its virulence.3 Our lab aims to address this by overexpressing and purifying recombinant Aft1, Grx3/4, and Bol2, so that the structure of Aft1 and its interactions with target genes and potential binding partners may be characterized. Aft1 involvement in iron regulation in C. glabrata is essential for the virulence of this pathogen,8 so these findings will improve understanding of how iron regulation impacts human susceptibility to fungal infections and have implications for the development of prophylactic or therapeutic treatments for fungal infections.
Methods
Plasmid Construction
The BOL2 gene was amplified from 10 ng, 25 ng, and 50 ng of C. glabrata genomic DNA (ATCC, Manassas, VA) by polymerase chain reaction (PCR) using forward and reverse primers (Table 1, Eurofins Genomics, Louisville, KY). To allow for ligation of the BOL2 sequence into the vector backbones, the forward primer incorporated a 5’ XhoI restriction enzyme site, and the reverse primer incorporated a 3’ PacI site. PCR reactions included 10 µL 5X Q5 Reaction Buffer, 10 mM dNTPs, 10 µM forward primer, 10 µM reverse primer, 0.5 µL Q5 High-Fidelity DNA Polymerase, 10 µL 5X Q5 High GC Enhancer, and nuclease-free water to 25 µL. PCR was performed with 30 cycles of melting temperature 98° C, annealing temperature 57.3 °C, and extension temperature 72 °C. Due to the 25 s/kb amplification rate of Q5 High-Fidelity DNA Polymerase and 297-bp PCR product, extension temperature was programmed for 25 s.
Table 1: Primers used for amplifying BOL2 from C. glabrata genome and cloning into E. coli expression vectors pRSFDuet-1 and pRSFDuet-1-Grx4.
|
Forward Primer |
ctgacgtcggtaccctcgagATGATTACTGAAGAACATCTG |
|
Reverse Primer |
ggcagcagcctaggttaattaaTTAAATTACTATCTTTGACCACTC |
*Lower-case nucleotides align with the pRSFDuet-1 and pRSFDuet-1-Grx4 vector backbones.
** Upper-case nucleotides align with the BOL2 gene.
*** Restriction sites used for cloning BOL2 are in bold.
PCR products were incubated with the restriction enzyme DpnI for 1 h at 37 °C for clean-up before confirmation of the products via agarose gel electrophoresis. The remaining products were combined for purification with the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI) protocol. The purified BOL2 fragment was cloned into pRSFDuet-1 (Novagen, Madison, WI) and pRSFDuet-1-Grx4 (E. Talib, C. Outten lab unpublished data) to generate pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2, respectively, via double digestion with XhoI and PacI restriction enzymes (Thermo Fisher Scientific, Waltham, MA) in CutSmart NEBuffer (New England Biolabs, Ipswich, MA). The pRSFDuet-1 and pRSFDuet-1-Grx4 vectors (1 µg) were incubated with the enzymes at 37 °C for 1 h to generate overlapping ends for annealing with the BOL2 gene; the reactions were further incubated at 65 °C for 20 min to inactivate the enzymes. The Gibson Assembly (GA) protocol (New England Biolabs) was used to ligate the Bol2 insert into the digested pRSFDuet-1 and pRSFDuet-1-Grx4 backbones through incubation for 15 or 30 min at 50 °C. The assembled constructs were confirmed with 1% agarose gel electrophoresis.
The pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2 plasmids were transformed into Escherichia coli DH5α cells on LB with 60 mg/mL kanamycin (Kan) and incubated at 37 °C overnight. Single colonies from each transformation plate were grown overnight in 5 mL LB-Kan media to provide four cultures of each construct. Plasmid DNA was isolated from the cells using the GeneJET Plasmid Miniprep kit (Thermo Fisher Scientific). To confirm assembly with double digestion, the pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2 DNA samples as well as corresponding vector backbone (5 µg) were incubated with restriction enzymes BspHI and SacI-HF at 37 °C for 1 h. The plasmid DNA samples were compared to the corresponding vector control by agarose gel electrophoresis. Sample colonies with the correct digestion pattern were sent for confirmation via DNA sequencing using DuetUP2 and T7-Term sequencing primers (Genewiz, Research Triangle Park, NC). Two of the confirmed pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2 DNA plasmid samples were transformed into E. coli BL21(DE3) cells and plated on LB-Kan.
Determination of Optimal Conditions for Small-Scale Overexpression
Single colonies were picked for pRSFDuet-1-Bol2/BL21(DE3) and pRSFDuet-1-Grx4-Bol2/BL21(DE3), The colonies were inoculated in small-scale cultures of LB with 60 mg/mL Kan and grown at 37 °C. The concentrations of the cells from each colony were checked by reading the absorbance at 600 nm (OD600). After 5 h of unexpected slow growth among colonies, each culture was streaked on new LB-Kan plates and incubated overnight at 37 °C for another attempt at culturing. One colony from each plate was inoculated in LB-Kan media and grown at 37 °C. An aliquot of non-induced cells was harvested prior to induction. After the OD600 for each culture reached 0.6, cultures were induced with 0.25 mM isopropyl β-D-thiogalactoside (IPTG) at 20 °C or 30 °C for either 4 h or overnight. One aliquot of the induced cells was harvested, and a second was treated with a bacterial protein extraction detergent (B-PER, Thermo Fisher Scientific) and centrifuged to extract the presumably soluble recombinant proteins from the cells and into the supernatant. Each non-induced, induced, and supernatant sample was evaluated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Results and Discussion
Plasmid Construction
Amplification and Ligation of Insert
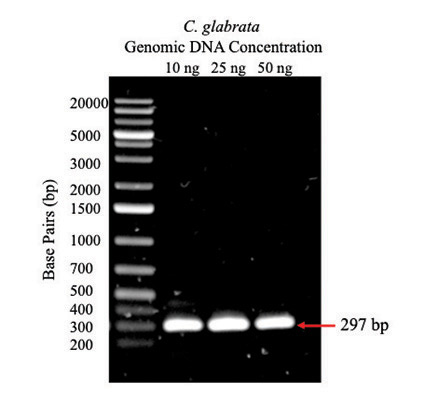
Figure 1: PCR amplification of BOL2 from C. glabrata genomic DNA. Agarose gel (1%) with 5-µL samples of PCR products from reactions containing 10 ng, 25 ng, and 50 ng C. glabrata genomic DNA; arrow indicates expected size of BOL2 fragment. The first lane is a DNA ladder with fragments of known size indicated to the left.
The BOL2 gene was amplified from C. glabrata genomic DNA using PCR. Agarose gel electrophoresis (Figure 1) shows bands at the expected size of 297 bp indicating that amplification of BOL2 was successful for each concentration of genomic DNA.
GA was utilized to ligate the BOL2 insert into the pRSFDuet-1 and pRSFDuet-1-Grx4 vector backbones forming expression plasmids for transformation into host cells. PCR products were digested with the restriction enzyme DpnI to remove methylated template DNA then further purified to isolate BOL2. The resulting fragment was high quality DNA with little RNA contamination as indicated by a 260 nm/280 nm reading of 1.86.
Successful incorporation of restriction enzyme sites into BOL2 and double digestion of vector backbones with XhoI and PacI allowed for ligation at the overlapping ends. Transformation of plasmids into E. coli DH5α resulted in amplification of recombinant DNA for Miniprep isolation.
Confirmation of Assembly with Vector Backbones
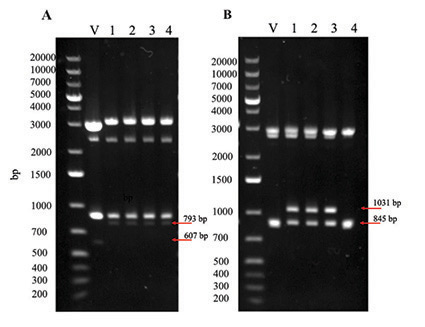
Figure 2: BspHI and SacI-HF double digestion for confirmation of pRSFDuet-1-Bol2 (A) and pRSFDuet-1-Grx4-Bol2 (B). Agarose gel (1%) showing successful double digestion of the plasmids and their respective vector backbone as a control. V = pRSFDuet-1 vector (A) or pRSFDuet-1-Grx4 vector (B). Numbers 1-4 indicate individual colonies from which plasmid DNA was isolated.
Assembly of complete expression vectors pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2 was confirmed by double digestion with BspHI and SacI-HF. Agarose gel electrophoresis for restriction digested pRSFDuet-1-Bol2 (Figure 2A) shows 793-bp bands for all colonies indicating successful ligation of BOL2 into the pRSFDuet-1 vector; the pRSFDuet-1 vector is missing the BOL2 gene as indicated by the shorter 607-bp band. Agarose gel electrophoresis for restriction digested pRSFDuet-1-Grx4-Bol2 (Figure 2B) shows 1031-bp bands for colonies 1-3 indicating successful ligation of BOL2 into the pRSFDuet-1-Grx4 vector; the pRSFDuet-Grx4 vector is missing the BOL2 gene as indicated by the shorter 845-bp band. The top bands (Figure 2) are undigested plasmid DNA further illustrating that the controls lack the BOL2 insert and are shorter. Constructs were confirmed by DNA sequencing before transformation into E. coli BL21(DE3) for protein expression.
Determination of Optimal Conditions for Small-Scale Overexpression
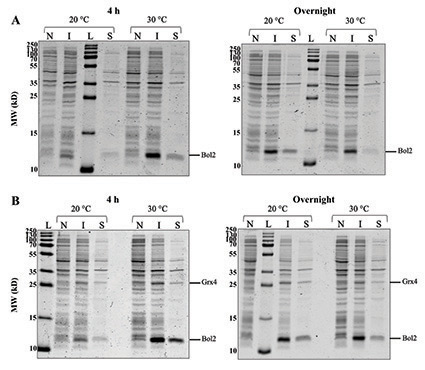
Figure 3: (A) Expression of recombinant Bol2 (expected MW = 9.5 kD). SDS-PAGE gels comparing Bol2 protein expression after induction with 0.25 mM IPTG and growth for two intervals at two temperatures. (B) Co-expression of Bol2 and Grx4 (expected MW = 29.8 kD). SDS-PAGE gels comparing Bol2 and Grx4 co-expression with same induction and growth conditions as (A). The sizes of the molecular weight ladder standards (MW; kD) are listed to the left of each gel. L = ladder, N = non-induced cells, I = induced cells, and S = supernatant.
Small-scale cultures inoculated with pRSFDuet-1-Bol2 and pRSFDuet-1-Grx4-Bol2 in the E. coli BL21(DE3) host strain were induced with 0.25 mM IPTG and grown for 4 h or overnight at 20 °C or 30 °C. Induction with 0.25 mM IPTG was based on the optimal conditions found by our lab during expression studies of Aft1 from C. glabrata (E. Talib, C. Outten lab unpublished data). Growth at 30 °C rather than the optimal E. coli growth temperature (37 °C) has been recommended for expression of soluble proteins in pET Vector Systems to avoid accumulation as inclusions bodies.9-10 Growth at 20 °C and accommodating with overnight incubation has been also been recommended for soluble protein expression.9-10 While protein degradation during the overnight condition is possible, these suggested conditions of lower temperature and prolonged growth may allow for better folding of soluble proteins and have been utilized for successful expression of recombinant Aft2, Bol2, and Grx3/4 from S. cerevisiae.4-6,9-11
SDS-PAGE gel electrophoresis of cells harvested from cultures of pRSFDuet-1-Bol2/BL21(DE3) and pRSFDuet-1-Grx4-Bol2/BL21(DE3) (Figure 3) depicts a band at ~11 kD in induced samples that is absent in non-induced samples indicating that Bol2 was successfully expressed after induction with IPTG both alone and in the presence of Grx4. The expected MW of Bol2 is 9.5 kD; however, on our gels Bol2 has consistently run at ~11 kD (Figure 3). This anomalous migration may be due to post-translational modifications of the protein or acidic residues that interfere in SDS binding during SDS-PAGE. Of note, Bol2 orthologs from other organisms also run at a higher MW than expected.5,6 Regardless of this slow migration tendency, the expression pattern between non-induced and induced conditions strongly suggests that this is the correct band. Protein mass spectrometry and size exclusion chromatography will be used in the future to assess the size of Bol2 from C. glabrata more accurately.
The 11-kD induced band for 4 h of growth at 30 °C (Figure 3A) appears sharper and more intense than the other bands of interest supporting that the optimal expression conditions tested for Bol2 alone were induction with 0.25 mM IPTG followed by growth for 4 h at 30 °C. SDS-PAGE gel electrophoresis of cells harvested from cultures of pRSFDuet-1-Grx4-Bol2/BL21(DE3) (Figure 3B) depicts a band at ~26 kD that is unique to the induced samples for the complexed construct and absent in non-induced samples indicating that Grx4 was successfully expressed in the presence of Bol2 after induction with IPTG. The 11-kD and 26-kD induced bands for 4 h of growth at 30 °C (Figure 3B) both appear sharper and more intense than the other bands of interest indicating that the optimal conditions tested for Bol2 and Grx4 co-expression were also induction with 0.25 mM IPTG followed by growth for 4 h at 30 °C. The intensity of the 11-kD band (Figure 3) is consistent for Bol2 alone and in the presence of Grx4 suggesting that Grx4 co-expression does not hinder Bol2 expression.
In the future, we will apply these conditions to large-scale expression studies for Bol2 and Grx4 and potential binding partners from C. glabrata. We will also confirm the identity of these proteins (Figure 3) with mass spectroscopy as well as purify them following the protocol previously described for the S. cerevisiae homologs.5
Conclusion
Our results show that the BolA-like protein Bol2 from C. glabrata was successfully recombinantly overexpressed in E. coli both alone and in the presence of the monothiol glutaredoxin Grx4. The optimal conditions for expression of Bol2 alone and co-expressed with Grx4 were found to be induction with 0.25 mM IPTG followed by incubation for 4 h at 30 °C. Bol2 and Grx4 will need to be purified for use in future interaction studies of these binding partners and the iron-responsive transcription factor Aft1 from C. glabrata. UV-visible absorption, circular dichroism, and electron paramagnetic resonance spectroscopies will be utilized to study the Fe-S cluster binding characteristics of Grx4 and Bol2 in the presence or absence of Aft1. Delineating these characteristics will further elucidate the role of Fe-S clusters in iron regulation in C. glabrata and indicate how regulation in this fungal pathogen compares to that of the non-pathogenic yeast S. cerevisiae.
About the Author
 Jade Ikahihifo-Bender
Jade Ikahihifo-Bender
I am a senior at the University of South Carolina who will graduate in May 2021 with a B.S. in Biochemistry & Molecular Biology and a minor in Psychology. After moving from Anchorage, Alaska, to Myrtle Beach, South Carolina, I have found immense support for my studies and research project at the University of South Carolina as a Carolina Scholar and recipient of the Science Undergraduate Research Fellowship grant and Magellan Scholar award. This project appealed to me because of its implications for the development of medications and therapies for iron regulation disorders as well as fungal infections. My research experience at USC encouraged me to pursue an internship at the National Institute of Allergy & Infectious Diseases where I worked in the laboratory of Dr. Jeffrey Cohen and expanded my interest in the implications of research. After graduation, I hope to work in clinical research or healthcare manufacturing to learn more about the complexities of healthcare before applying to medical school.
This work is an extension of a project that was previously presented for independent study, Discover USC 2019, and Graduation with Leadership Distinction in Research. It will also be included in my Honors Senior Thesis and Discover USC 2021 presentation. I would like to extend my gratitude to Dr. Caryn Outten in the Department of Chemistry & Biochemistry for the opportunity to work in her lab for over five semesters and for her guidance. Mrs. Evan Talib has been instrumental in my growth and project progression with her instruction and unwavering encouragement. I would also like to acknowledge the C. Outten lab group as a whole for their support.
This work was funded by the National Institute of General Medical Sciences R01 (Grant GM100069 to CEO), SC Research Foundation, SCHC Science Undergraduate Research Fellowship grant, and UofSC Magellan Scholar award.
References
- Wallace DF (2016) The regulation of iron absorption and homeostasis. Clin Biochem Rev 37(2):51-62.
- Himmelfarb J (2007) Iron regulation. JASN 18(2):379-381.
- Devaux F, Thiébaut A (2019) The regulation of iron homeostasis in the fungal human pathogen Candida glabrata. Microbiology 165(10):1041-1060.
- Poor CB, Wegner SV, Li H, Dlouhy AC, Scheurmann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C (2014) Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. PNAS 111(11):4043-4048.
- Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, and Outten CE (2009) The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48(40):9569-9581.
- Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE (2011) Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem 286(1):867-876.
- Ojeda L, Keller G, Mühlenhoff U, Rutherford JC, Lill R, Winge DR (2006) Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem 281(26):17661-17669.
- Gerwien F, Safyan A, Wisgott S, Hille F, Kaemmer P, Linde J, Brunke S, Kasper L, Hube B (2016) A novel hybrid iron regulation network combines features from pathogenic and nonpathogenic yeasts. ASM 7(5):1-16.
- Novagen, Inc. (1999) pET system manual. United States and Canada. TB055 8th Edition 02/99. 8th ed. Available from: http://research.fhcrc.org/content/dam/stripe/hahn/methods/biochem/pet.pdf .
- Novagen, Inc. (2006) pET system manual. United States and Canada. TB055 11th Edition 01/06. 11th ed. Available from: http://kirschner.med.harvard.edu/files/protocols/Novagen_petsystem.pdf .
- Li H, Outten CE (2019) The conserved CDC motif in the yeast iron regulator Aft2 mediates iron–sulfur cluster exchange and protein–protein interactions with Grx3 and Bol2. J Biol Inorg. 48(40):9569-9581
